Quant II Recitation
Drew Dimmery
January 30, 2015
What is this?
- This presentation created using knitr + pandoc + reveal.js
- I have created a recitation repository on github: https://github.com/ddimmery/Quant-II-Recitation
- The recitation website will also be on github, at http://ddimmery.github.io/Quant-II-Recitation
- This presentation is available on the course website, as is a handout version in pdf.
The raw .Rmd file is available in the repository. You can create a handout version of it with the following commands (assuming knitr and pandoc are installed), first in R:
require(knitr) knit(presentation.Rmd)And then on the command line:
pandoc presentation.md -o handout.pdf
Today’s Topic
- Sharing is caring.
- If you don’t share your research (+ code and data) then what’s the point of doing it?
- Today we will be discussing some necessary mechanics.
- R and how to bend it to your will
- Producing high quality tables / plots
- If there’s any time left, we can talk a little more about identification
Sharing
- All homework will be submitted to me (preferably in hard copy)
- All replication code will be posted to a gist on github, and submitted to me via email.
- gist.github.com
- Code should have a header with relevant information (name, date created, date modified, input files, output files, etc)
- Code should be well commented
- If you’d prefer to submit homework as some sort of knitr document, that is also fine. Just submit the
.Rmdfile.
- All tables and plots should be of very high quality.
- Yes, this will take a non-trivial amount of time.
Workflow
When things break
- Documentation - Ex:
?lm - CRAN (Reference manuals, vignettes, etc) - Ex: http://cran.r-project.org/web/packages/AER/index.html
- JSS - Ex: http://www.jstatsoft.org/v27/i02
- Stack Overflow - http://stackoverflow.com/questions/tagged/r
- Listservs - http://www.r-project.org/mail.html
Resources
- The Art of R Programming - N. Matloff
- Modern Applied Statistics with S - W. Venables and B. Ripley
- Advanced R Programming - forthcoming, H. Wickham
- The R Inferno - P. Burns
- Rdataviz - a talk by P. Barberá on ggplot2
- Basic Intro to R - also by P. Barberá
Reading R Documentation
CRAN documentation
JSS
Confusing Things
- At the prompt, > means you’re good to go, + means a parenthesis or bracket is open.
- Case sensitive
- Use / in path names. Not \.
- R uses variables – there is no “sheet” here, like in Stata
- R is a programming language
- More on errors later!
Using Third-party Code
- Relevant commands are:
install.packagesandlibrary Find the appropriate packages and commands with Google and via searching in R:
?covariance ??covariance install.packages("sandwich") library("sandwich") ?vcovHC
Data types
- Character - strings
- Double / Numeric - numbers
- Logical - true/false
- Factor - unordered categorical variables
- Objects - its complicated
Character
str <- "This is a string"paste("This", "is", "a", "string", sep = " ")## [1] "This is a string"as.character(99)## [1] "99"class(str)## [1] "character"Numeric
num <- 99.867
class(num)## [1] "numeric"round(num, digits = 2)## [1] 99.87round(str, digits = 2)## Error: non-numeric argument to mathematical functionpi## [1] 3.142exp(1)## [1] 2.718sin,exp,log,factorial,choose,BesselJ, etc
Logical
- The logical type allows us to make statements about truth
2 == 4## [1] FALSEclass(2 == 4)## [1] "logical"str != num## [1] TRUE"34" == 34## [1] TRUE==,!=,>,<,>=,<=,!,&,|,any,all, etc
Objects
- Many functions will return objects rather than a single datatype.
X <- 1:100
Y <- rnorm(100, X)
out.lm <- lm(Y ~ X)
class(out.lm)## [1] "lm"- Objects can have other data embedded inside them
out.lm$rank## [1] 2class(out.lm$rank)## [1] "integer"Data Structures
There are other ways to hold data, though:
- Vectors
- Lists
- Matrices
- Dataframes
Vectors
- Almost everything in R is a vector.
as.vector(4)## [1] 44## [1] 4- We can combine vectors with
c:
vec <- c("a", "b", "c")
vec## [1] "a" "b" "c"c(2, 3, vec)## [1] "2" "3" "a" "b" "c"Vectors (cont.)
- Sometimes R does some weird stuff:
c(1, 2, 3, 4) + c(1, 2)## [1] 2 4 4 6- It “recycles” the shorter vector:
c(1, 2, 3, 4) + c(1, 2, 1, 2)## [1] 2 4 4 6c(1, 2, 3, 4) + c(1, 2, 3)## Warning: longer object length is not a multiple of shorter object length## [1] 2 4 6 5More Vectors
- We can index vectors in several ways
vec[1]## [1] "a"names(vec) <- c("first", "second", "third")
vec## first second third
## "a" "b" "c"vec["first"]## first
## "a"Missingness
vec[1] <- NA
vec## first second third
## NA "b" "c"is.na(vec)## first second third
## TRUE FALSE FALSEvec[!is.na(vec)] # vec[complete.cases(vec)]## second third
## "b" "c"Lists
- Lists are similar to vectors, but they allow for arbitrary mixing of types and lengths.
listie <- list(first = vec, second = num)
listie## $first
## first second third
## NA "b" "c"
##
## $second
## [1] 99.87listie[[1]]## first second third
## NA "b" "c"listie$first## first second third
## NA "b" "c"Matrices
- \[A = \begin{pmatrix}1 & 3\\ 2 & 4\end{pmatrix}\]
- \(A_{ij}\)
- \(A_{1,2} = 3\)
- \(A_{1,\cdot} = (1,3)\)
A <- matrix(c(1, 2, 3, 4), nrow = 2, ncol = 2)
A## [,1] [,2]
## [1,] 1 3
## [2,] 2 4A[1, 2]## [1] 3A[1, ]## [1] 1 3Matrix Operations
- Its very easy to manipulate matrices:
solve(A) #A^{-1}## [,1] [,2]
## [1,] -2 1.5
## [2,] 1 -0.510 * A## [,1] [,2]
## [1,] 10 30
## [2,] 20 40B <- diag(c(1, 2))
B## [,1] [,2]
## [1,] 1 0
## [2,] 0 2A %*% B## [,1] [,2]
## [1,] 1 6
## [2,] 2 8More Matrix Ops.
A %*% diag(3)## Error: non-conformable argumentst(A) # A'## [,1] [,2]
## [1,] 1 2
## [2,] 3 4rbind(A, B)## [,1] [,2]
## [1,] 1 3
## [2,] 2 4
## [3,] 1 0
## [4,] 0 2cbind(A, B)## [,1] [,2] [,3] [,4]
## [1,] 1 3 1 0
## [2,] 2 4 0 2c(1, 2, 3) %x% c(1, 1) # Kronecker Product## [1] 1 1 2 2 3 3Naming Things
rownames(A)## NULLrownames(A) <- c("a", "b")
colnames(A) <- c("c", "d")
A## c d
## a 1 3
## b 2 4A[, "d"]## a b
## 3 4- Matrices are vectors:
A[3]## [1] 3Dataframes
The workhorse
Basically just a matrix that allows mixing of types.
data(iris)
head(iris)## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosaControl Flow
- loops
- if/else
- functions
- useful stock functions to know
Loops
- for loops - just a way to say “do this for each element of the index”
- “this” is defined in what follows the “for” expression
for (i in 1:5) {
cat(i * 10, " ")
}## 10 20 30 40 50for (i in 1:length(vec)) {
cat(vec[i], " ")
}## NA b cfor (i in vec) {
cat(i, " ")
}## NA b cIf/Else
if (vec[2] == "b") print("Hello World!")## [1] "Hello World!"if (vec[3] == "a") {
print("Hello World!")
} else {
print("!dlroW olleH")
}## [1] "!dlroW olleH"Vectorized If/Else
- Conditional execution on each element of a vector
vec <- letters[1:3]
new <- vector(length = length(vec))
for (i in 1:length(vec)) {
if (vec[i] == "b") {
new[i] <- 13
} else {
new[i] <- 0
}
}
new## [1] 0 13 0new <- ifelse(vec == "b", 13, 0)
new## [1] 0 13 0Functions
- \(f : X \to Y\)
- Functions in R are largely the same. (“Pure functions”)
add3 <- function(X) {
return(X + 3)
}
add3(2)## [1] 5makeGroups <- function(groups, members = 1) {
return((1:groups) %x% rep(1, members))
}
makeGroups(5)## [1] 1 2 3 4 5makeGroups(5, 2)## [1] 1 1 2 2 3 3 4 4 5 5Useful Functions
Note: Most functions don’t do complete case analysis by default (usually option na.rm=TRUE)
print,cat,paste,with,length,sort,order,unique,rep,nrow,ncol,complete.cases,subset,merge,mean,sum,sd,var,lag,lm,model.matrix,coef,vcov,residuals,vcovHC(fromsandwich),ivreg(fromAER),countrycode(fromcountrycode),summary,pdf,plot, Tools fromplm, and many more.
Distributional Functions
?Distributions- They have a consistent naming scheme.
rnorm,dnorm,qnorm,pnormrdist- generate random variable from distddist- density function of distqdist- quantile function of distpdist- distribution function of dist- look at documentation for parameterization
rnorm(16)## [1] 0.03219 -0.89729 -0.28782 0.86993 -1.21937 1.47985 0.38488
## [8] 0.28917 -1.66721 0.23155 1.63280 0.84529 -0.87946 -0.22374
## [15] 1.35861 -0.61532Robust SEs
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 | |
Multiple Dispactch
- Sometimes functions will behave differently based on context:
summary(vec)## Length Class Mode
## 3 character charactersummary(c(1, 2, 3, 4))## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1.00 1.75 2.50 2.50 3.25 4.00summary(iris[, 1:4])## Sepal.Length Sepal.Width Petal.Length Petal.Width
## Min. :4.30 Min. :2.00 Min. :1.00 Min. :0.1
## 1st Qu.:5.10 1st Qu.:2.80 1st Qu.:1.60 1st Qu.:0.3
## Median :5.80 Median :3.00 Median :4.35 Median :1.3
## Mean :5.84 Mean :3.06 Mean :3.76 Mean :1.2
## 3rd Qu.:6.40 3rd Qu.:3.30 3rd Qu.:5.10 3rd Qu.:1.8
## Max. :7.90 Max. :4.40 Max. :6.90 Max. :2.5- Why?
?summary?summary.lm
The *apply family
- These functions allow one to efficiently perform a large number of actions on data.
apply- performs actions on the rows or columns of a matrix/array (1 for rows, 2 for columns, 3 for ??)sapply- performs actions on every element of a vectortapply- performs actions on a vector by groupreplicate- performs the same action a given number of times
apply
A## c d
## a 1 3
## b 2 4apply(A, 1, sum)## a b
## 4 6apply(A, 2, mean)## c d
## 1.5 3.5sapply
vec## [1] "a" "b" "c"sapply(vec, function(x) paste0(x, ".vec"))## a b c
## "a.vec" "b.vec" "c.vec"- Can be accomplished more simply with:
paste0(vec, ".vec")## [1] "a.vec" "b.vec" "c.vec"Why?
replicateis basically justsapply(1:N,funct)wherefunctnever uses the index.
tapply
tapply(1:10, makeGroups(5, 2), mean)## 1 2 3 4 5
## 1.5 3.5 5.5 7.5 9.5Working With Data
- Input
- Output
Input
setwd("~/github/Quant II Recitation/2014-01-31/")
dir()## [1] "apsrtable.png" "figure" "handout.pdf"
## [4] "iris.csv" "presentation.html" "presentation.md"
## [7] "presentation.Rmd" "stargazer.png"iris <- read.csv("iris.csv")If data is, for instance, a Stata .dta file, use
read.dtafrom theforeignpackage.Useful options for reading data:
sep,na.strings,stringsAsFactorsFor different formats, Google it.
Simulate some Data
set.seed(1023) # Important for replication
X <- rnorm(1000, 0, 5)
Y <- sin(5 * X) * exp(abs(X)) + rnorm(1000)
dat <- data.frame(X, Y)
plot(X, Y, xlim = c(0, 5), ylim = c(-50, 50))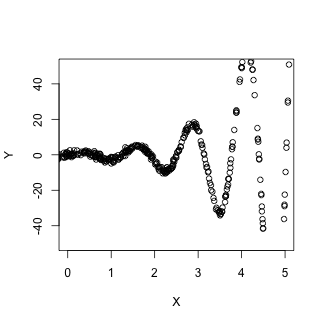
Regression Output
dat.lm <- lm(Y ~ X, data = dat)
dat.lm##
## Call:
## lm(formula = Y ~ X, data = dat)
##
## Coefficients:
## (Intercept) X
## -216634 183687Regression Output
summary(dat.lm)##
## Call:
## lm(formula = Y ~ X, data = dat)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.10e+08 -4.19e+05 2.01e+05 8.17e+05 9.08e+06
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -216634 212126 -1.02 0.31
## X 183687 43470 4.23 2.6e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 6710000 on 998 degrees of freedom
## Multiple R-squared: 0.0176, Adjusted R-squared: 0.0166
## F-statistic: 17.9 on 1 and 998 DF, p-value: 2.6e-05Pretty Output
- How do we get LaTeX output?
- The
xtablepackage:
require(xtable)## Loading required package: xtablextable(dat.lm)## % latex table generated in R 3.0.2 by xtable 1.7-1 package
## % Tue Jan 28 23:38:40 2014
## \begin{table}[ht]
## \centering
## \begin{tabular}{rrrrr}
## \hline
## & Estimate & Std. Error & t value & Pr($>$$|$t$|$) \\
## \hline
## (Intercept) & -216633.6722 & 212125.4622 & -1.02 & 0.3074 \\
## X & 183687.1735 & 43469.5839 & 4.23 & 0.0000 \\
## \hline
## \end{tabular}
## \end{table}xtable
xtableworks on any sort of matrix
xtable(A)## % latex table generated in R 3.0.2 by xtable 1.7-1 package
## % Tue Jan 28 23:38:40 2014
## \begin{table}[ht]
## \centering
## \begin{tabular}{rrr}
## \hline
## & c & d \\
## \hline
## a & 1.00 & 3.00 \\
## b & 2.00 & 4.00 \\
## \hline
## \end{tabular}
## \end{table}xtable
- This is what
xtabledoes with thelmobject:
class(summary(dat.lm)$coefficients)## [1] "matrix"xtable(summary(dat.lm)$coefficients)## % latex table generated in R 3.0.2 by xtable 1.7-1 package
## % Tue Jan 28 23:38:40 2014
## \begin{table}[ht]
## \centering
## \begin{tabular}{rrrrr}
## \hline
## & Estimate & Std. Error & t value & Pr($>$$|$t$|$) \\
## \hline
## (Intercept) & -216633.67 & 212125.46 & -1.02 & 0.31 \\
## X & 183687.17 & 43469.58 & 4.23 & 0.00 \\
## \hline
## \end{tabular}
## \end{table}- Note that this is the same as the output from
xtable(dat.lm)
Pretty it up
- Now let’s make some changes to what
xtablespits out:
print(xtable(dat.lm, digits = 1), booktabs = TRUE)## % latex table generated in R 3.0.2 by xtable 1.7-1 package
## % Tue Jan 28 23:38:40 2014
## \begin{table}[ht]
## \centering
## \begin{tabular}{rrrrr}
## \toprule
## & Estimate & Std. Error & t value & Pr($>$$|$t$|$) \\
## \midrule
## (Intercept) & -216633.7 & 212125.5 & -1.0 & 0.3 \\
## X & 183687.2 & 43469.6 & 4.2 & 0.0 \\
## \bottomrule
## \end{tabular}
## \end{table}- Many more options, see
?xtableand?print.xtable
apsrtable
- Read the documentation - there are many options.
require(apsrtable)## Loading required package: apsrtabledat.lm2 <- lm(Y ~ X + 0, data = dat)
apsrtable(dat.lm, dat.lm2)## Note: no visible binding for global variable 'se'
## Note: no visible binding for global variable 'se'
## Note: no visible binding for global variable 'nmodels'
## Note: no visible binding for global variable 'lev'## \begin{table}[!ht]
## \caption{}
## \label{}
## \begin{tabular}{ l D{.}{.}{2}D{.}{.}{2} }
## \hline
## & \multicolumn{ 1 }{ c }{ Model 1 } & \multicolumn{ 1 }{ c }{ Model 2 } \\ \hline
## % & Model 1 & Model 2 \\
## (Intercept) & -216633.67 & \\
## & (212125.46) & \\
## X & 183687.17 ^* & 182921.71 ^*\\
## & (43469.58) & (43464.06) \\
## $N$ & 1000 & 1000 \\
## $R^2$ & 0.02 & 0.02 \\
## adj. $R^2$ & 0.02 & 0.02 \\
## Resid. sd & 6706998.86 & 6707143.05 \\ \hline
## \multicolumn{3}{l}{\footnotesize{Standard errors in parentheses}}\\
## \multicolumn{3}{l}{\footnotesize{$^*$ indicates significance at $p< 0.05 $}}
## \end{tabular}
## \end{table}apsrtable
library(png)
library(grid)
img <- readPNG("apsrtable.png")
grid.raster(img)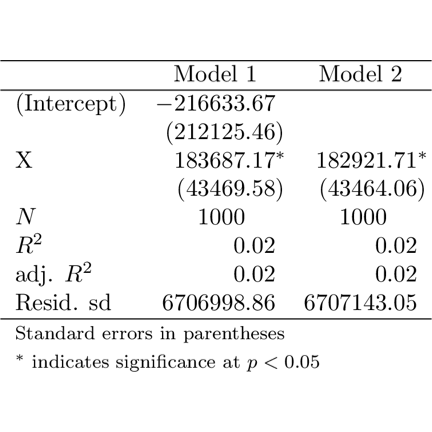
stargazer
require(stargazer)## Loading required package: stargazer
##
## Please cite as:
##
## Hlavac, Marek (2013). stargazer: LaTeX code and ASCII text for well-formatted regression and summary statistics tables.
## R package version 4.5.3. http://CRAN.R-project.org/package=stargazerstargazer(dat.lm, dat.lm2)##
## % Table created by stargazer v.4.5.3 by Marek Hlavac, Harvard University. E-mail: hlavac at fas.harvard.edu
## % Date and time: Tue, Jan 28, 2014 - 23:38:48
## \begin{table}[!htbp] \centering
## \caption{}
## \label{}
## \begin{tabular}{@{\extracolsep{5pt}}lcc}
## \\[-1.8ex]\hline
## \hline \\[-1.8ex]
## & \multicolumn{2}{c}{\textit{Dependent variable:}} \\
## \cline{2-3}
## \\[-1.8ex] & \multicolumn{2}{c}{Y} \\
## \\[-1.8ex] & (1) & (2)\\
## \hline \\[-1.8ex]
## X & 183,687.000$^{***}$ & 182,922.000$^{***}$ \\
## & (43,470.000) & (43,464.000) \\
## & & \\
## Constant & $-$216,634.000 & \\
## & (212,125.000) & \\
## & & \\
## \hline \\[-1.8ex]
## Observations & 1,000 & 1,000 \\
## R$^{2}$ & 0.018 & 0.017 \\
## Adjusted R$^{2}$ & 0.017 & 0.016 \\
## Residual Std. Error & 6,706,999.000 (df = 998) & 6,707,143.000 (df = 999) \\
## F Statistic & 17.860$^{***}$ (df = 1; 998) & 17.710$^{***}$ (df = 1; 999) \\
## \hline
## \hline \\[-1.8ex]
## \textit{Note:} & \multicolumn{2}{r}{$^{*}$p$<$0.1; $^{**}$p$<$0.05; $^{***}$p$<$0.01} \\
## \normalsize
## \end{tabular}
## \end{table}stargazer
img <- readPNG("stargazer.png")
grid.raster(img)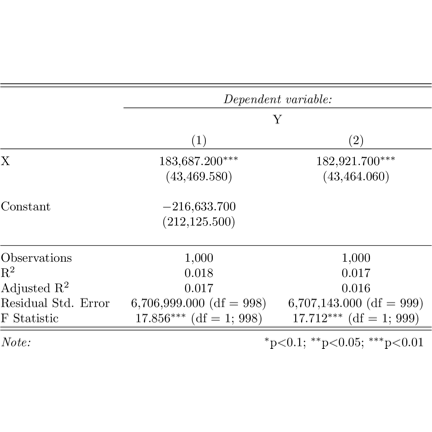
Both
Both packages are good (and can be supplemented with
xtablewhen it is easier)Get pretty close to what you want with these packages, and then tweak the LaTeX directly.
Plotting
- It’s all about coordinate pairs.
plot(x,y)plots the pairs of points inxandy- Notable options:
type- determines whether you plot points, lines or whatnotpch- determines plotting characterxlim- x limits of the plot (likewise fory)xlab- label on the x-axismain- main plot labelcol- color- A massive number of options. Read the docs.
- Some objects respond specially to
plot. Tryplot(dat.lm)
Tying it Together
x <- seq(-1, 1, 0.01)
y <- 3/4 * (1 - x^2)
plot(x, y, type = "l", xlab = "h", ylab = "weight")
Tying it Together
- \(W\) is an \(n\times p\) diagonal weighting matrix, \(h\) is a “bandwidth”.
- Diagonal entries are \(\frac{3}{4}\cdot(1-d^2)\cdot 1_{\{|d|\le 1\}}\) where \(d = \frac{X-c}{h}\)
- \(\hat{\beta}_c = (X'WX)^{-1}X'WY\)
- Covariance matrix is \(s^2(X'WX)^{-1}\)
loc.lin <- function(Y, X, c = 0, bw = sd(X)/2) {
d <- (X - c)/bw
W <- 3/4 * (1 - d^2) * (abs(d) < 1)
W <- diag(W)
X <- cbind(1, d)
b <- solve(t(X) %*% W %*% X) %*% t(X) %*% W %*% Y
sigma <- t(Y - X %*% b) %*% W %*% (Y - X %*% b)/(sum(diag(W) > 0) - 2)
sigma <- solve(t(X) %*% W %*% X) * c(sigma)
return(c(est = b[1], se = sqrt(diag(sigma))[1]))
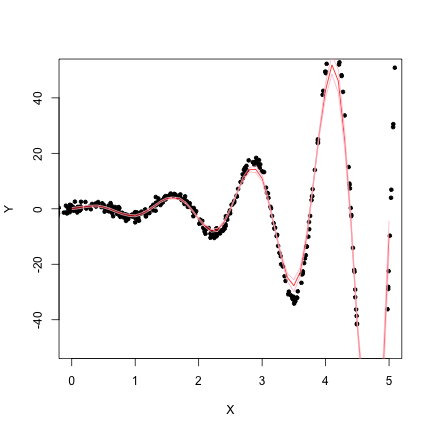
}Fit the Surface
X.est <- seq(0, 5, 0.1)
dat.llm <- sapply(X.est, function(x) loc.lin(Y, X, c = x, bw = 0.25))
plot(X, Y, xlim = c(0, 5), ylim = c(-50, 50), pch = 20)
lines(X.est, dat.llm[1, ], col = "red")
lines(X.est, dat.llm[1, ] + 1.96 * dat.llm[2, ], col = "pink")
lines(X.est, dat.llm[1, ] - 1.96 * dat.llm[2, ], col = "pink")